bioECS
The recent progress in molecular biology and in genomics, as well as new developments in terms of data-acquisition devices have allowed the complete sequencing of the human genome, and other species. Consequently, the databases which collect these genomic sequences have grown in an expontential fashion these last years. Among these databases the most used ones are the GenBank and the EMBL databases. The information contained in these databases has become central to the researchers in biology and is therefore important to develop effective bioinformatics tools to help them to efficiently access and analyse this information.
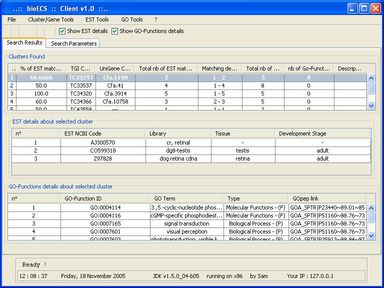
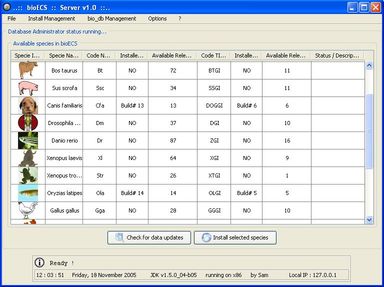
Within this context, the aim of the present work was to develop an ad hoc search engine to help biologists to access (collect, filter, sort, store locally) the data they need in an as easy and time-sparing way possible. The functional specification of the resulting software (called BioECS, for 'Biology Easy Cluster Search') and its user interface have been defined in collaboration with end-users from the 'Centre of Biomedical Integrative Genoproteomics' (CBIG) of the University of Liège. The study which has motivated the development of the software aims at studying genes related to the development of pancreas tissues in Zebra Fish (Danio Rerio), but the resulting software is general and can be used in the context of similar studies. The program provides a solution to biologists in order to isolate EST (Expressed Sequence Tags) clusters corresponding to their research target. A simple Internet surfing software makes this interaction intuitive. The software also provides facilities for saving query results and search criteria for later use. Our solution is based on cross references between several EST databases, namely TIGR and Unigene. The resulting software allows to filter out EST clusters by combining several criteria based on information about tissues, species, and development stages associated to the EST sequences that form the clusters. Each criterion is associated with a "percentage" threshold and the clusters that do not contain enough EST sequences satisfying the criterion are filtered out. The user interface allows to easily access all EST sequences forming the clusters and to browse known GO (Gene Ontology) functions associated to them.
The client/server software architecture is based on JAVA and MySQL. It comprises a 'server' program responsible for the web-based transactions and local caching of information from the public database servers. A 'client' program allows biologists to execute researches. These latter use the JDBC API (Sun MicroSystems) to allow the user to interact in a flexible way with the server, to obtain the selected data items and create reports.
This work was achieved by Benjamin Renwart during his final thesis (2004-2005).
Download Jar file and Source code
Documentation
Benjamin Renwart Final thesis (in french)